Porcine circovirus 2 (PCV-2) genetic variability under natural infection scenario reveals a complex network of viral quasispecies.
A study revealing the complex network of viral quasiespecies present under natural infection scenario of porcine circovirus 2 (PCV-2), was published recently by researchers from IRTA-CReSA in collaboration with a researcher from University of Padua (Italy).
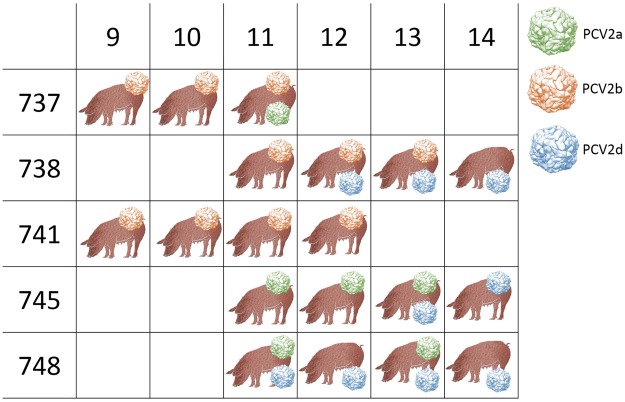
The main objective of this study was to analyze the within-host genetic variability of PCV-2 subpopulations (or quasiespecies, see box) to unravel the forces driving PCV-2 evolution. To accomplish this, a longitudinal weekly sampling was conducted on individual animals located in three farms after the first PCV-2 detection. The analysis of polymorphisms evaluated throughout the full PCV-2 genome demonstrated the presence of several single nucleotide polymorphisms (SNPs) especially in the genome region encoding for the capsid gene. The global haplotype reconstruction allowed inferring the virus transmission network over time, suggesting a relevant within-farm circulation. Evidences of co-infection and recombination involving multiple PCV-2 genotypes were found after mixing with pigs originating from other sources.
To understand the meaning of the term ‘quasispecies’, is useful to analyze its etymology. Quasiespecies are clouds of related elements that behave almost (quasi) like a single type of molecule (species)
The present study demonstrates the remarkable genetic variability of PCV-2 quasispecies that occur within the host, suggesting the role of the natural selection induced by the host immune response in driving PCV-2 evolution. Moreover, the effect of pig management in multiple genotype coinfections occurrence and recombination likelihood was demonstrated.
You can download the paper in the following link:
Correa-Fiz F, Franzo G, Llorens A, Segalés J, Kekarainen T Porcine circovirus 2 (PCV-2) genetic variability under natural infection scenario reveals a complex network of viral quasispecies. Scientific Reports. 2018
The author is part of the projects “Evaluation of animal models and protection against Glässer’s disease and the role of respiratory microbiota” (AGL2013-45662-R) funded by MINECO and PIGSs (Program for Innovative Global Prevention of Streptococcus suis) financed by the European Union (H2020-EU 727966), where the microbiota of pigs is explored in relation to different pathologies. This work was supported by the Boehringer Ingelheim European Animal Health PCV-2 award (2014).













