Gotta misfold’em all!

At IRTA-CReSA, we have been working on animal prion diseases for more than 20 years. Nowadays, almost no one remembers mad cows, but the truth is that these rare diseases remain a mystery and no one has yet been able to discover a treatment that works.
If you are a follower of this blog, you may remember that in previous projects we had focused on understanding why some species, such as dogs or horses, did not have prion diseases. Those projects gave interesting enough results but… why stick with just these two species?
At IRTA-CReSA we participate in the ZOOPRION project partially funded by the Spanish Ministry of Science and Innovation (PID2021-122201OB-C22), led by the group of Dr. Joaquín Castilla from CICbioGUNE, with whom we have collaborated closely for many years.
In the CICbioGUNE laboratories, they have developed and perfected a technique that will surely become revolutionary in the world of scientists who study prions: the PMSA or protein misfolding shaking amplification. This technique allows, in a very simple way, to misfold a solution of cellular prion protein in a test tube and transform it into infectious prions within a few hours. Mind-boggling if you think that it takes years for prions to appear in an animal. Thus, one of the objectives of this project has been to explore for the sequences of hundreds of cellular prion proteins from many different species, synthesize them in the laboratory and study their misfolding proneness.
In a funny analogy to PokéDex, the encyclopaedia of Pokémon species, the PrPdex website has been published. On this website, the project yields to the scientific community a huge amount of data on the structure and misfolding capacity of hundreds of mammal species. We invite you to visit it and find out what is misfolding potential of the prion protein of your favourite animal…. you’re sure to find it there, so far there are 725 species!
More information can be found in the recent publication in the journal Nature Communications:
Hasier Eraña, Cristina Sampedro-Torres-Quevedo, Jorge M. Charco, Carlos M. Díaz-Domínguez, Francesca Peccati, Maitena San-Juan-Ansoleaga, Enric Vidal, Nuno Gonçalves-Anjo, Miguel A. Pérez-Castro, Ezequiel González- Miranda, Patricia Piñeiro, Leire Fernández-Veiga, Josu Galarza-Ahumada, Eva Fernández-Muñoz, Guiomar Perez de Nanclares, Glenn Telling, Mariví Geijo, Gonzalo Jiménez-Osés & Joaquín Castilla
The potential of this tool is immense, it will allow us to determine which amino acid combinations of the prion protein are the most resistant to misfolding, or on the contrary, the most susceptible. With this information, it will be possible to design better tools to block the progression of prions and more efficient models for the study of new therapeutic strategies.
In addition, the index of misfolding proneness of each species is certainly a good predictor of its susceptibility.
For example, if we want to know whether the native cervid species of Catalonia would be susceptible to prions in the event of CWD (chronic wasting disease of cervids) we can look them up in the PrPdex and we will see that equally the fallow deer, the red deer and the roe deer have a misfolding potential index of 100! In other words, the synthetic prion protein of these species has been misfolded in 100% of the PMSA reactions that have been carried out in the laboratory. This is an indicator of the potential susceptibility of our cervids and explains why this disease is so difficult to control in North American cervid populations.
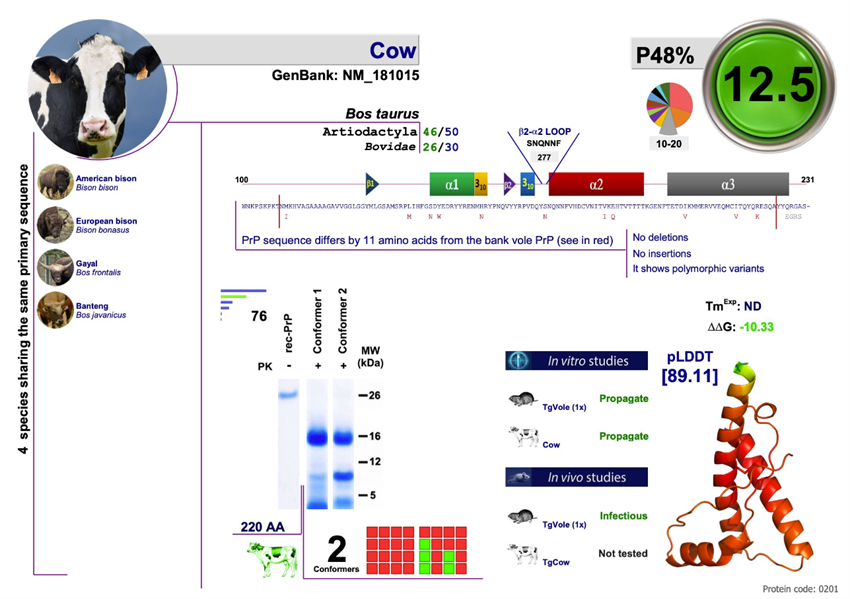
Example of a file from the PrPdex.com website, in this case of the bovine species.
Humans have an index of 30.4 and cows a 12.5… things could have been much worse!!
Currently, the website only includes the results of one polymorphism for each species (that is, a single variant of the prnp gene, of the many that can exist in the same species), but this information will be incorporated over time. It will certainly be key when it comes to refining the already existing genetic selection policies for prion-resistant livestock.
IRTA-CReSA’s contribution to this work has been to demonstrate, through bioassays in mouse models, that indeed some of these prions generated in test tubes behave as infectious agents, that is, they can infect and make a living being, such as mouse, sick with a prion disease. This step is key to extrapolate the in vitro results of the rest of the species and we can perform it thanks to having an animal house within the level 3 high biological containment unit (ICTS RLASB node). We are also studying the transmissibility of these synthetic prions to humans through bioassay in humanized transgenic mice.
The validation of this in vitro tool will make it possible to obtain information on the susceptibility to prions of hitherto unstudied species, saving the use of hundreds of experimental animals. It will also, therefore, represent an important advance in terms of reducing the use of experimental animals.














